Upload Data
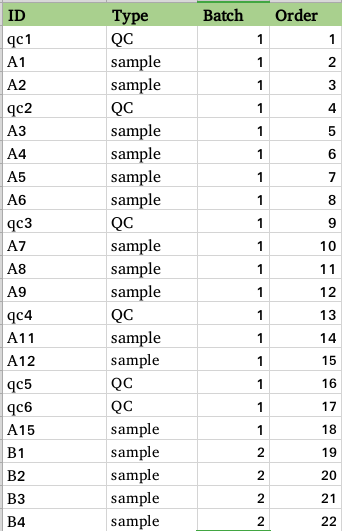
notes: Upload Omics Data's first column must be unique ;Group Design must have columns: ID,Type,Batch.
'ID' is the sample name contained in Omics Data; 'Type' is the grouping for sample matching; 'Batch' is a batch grouping of samples; 'Order' is the injection sequence.
Omics Data detail
notes: The table only displays the information of the first 100 rows and 100 columns of the upload data
Group Design detail
Note: Filtered missing values will be retained without being imputed.
Pre-process Omics file
notes: The table only displays the information of the first 100 rows and 100 columns of the Pre-process omics file
Please choose:
PCA plot
Please choose:
Download:
PVCA Barplot
PVCA Pieplot
Pearson correlation heatmap
Pearson correlation boxplot
Please choose:
UMAP plot
Please choose:
TSNE plot
Download:
Volcano plot
Differential barplot
Statistics Omics file
Note: SERRF method requires that samples should be at least 6 QCs in each batch.
After batch effect correction
notes: The table only displays the information of the first 100 rows and 100 columns of the after correct batch effect omics file
Combat correct batch effect
Combat has a very flexible EB framework for adjusting for additive, multiplicative, and exponential (when data have been log transformed) batch effects. Allow for the combination of multiple data sets and are robust to small or big sample sizes.
SERRF correct batch effect
A novel approach based on using quality control pool samples (QC). This method is called systematic error removal using random forest (SERRF) for eliminating the unwanted systematic variations in large sample sets.
Please choose:
PCA plot
Please choose:
Download:
PVCA Barplot
PVCA Pieplot
Pearson correlation heatmap
Pearson correlation boxplot
Please choose:
UMAP plot
Please choose:
TSNE plot
Download:
Volcano plot
Differential barplot
Statistics Omics file
Omics Batch Correct
Welcome !
Proteomics, metabolomics and lipomics based on mass spectrometry have become the key research methods. However, In the data collection process of large cohorts, batch effects due to non-biological influences are unavoidable. We compared different batch correction methods on different datasets and developed a pipeline software for evaluating and correcting omics data. The best correction method for mass spectrometry data omics is Combat. The process software evaluates the omics data and corrects the data after the specific batch effect. It is used to obtain reliable and real omics data and provide a research basis for large-scale omics. V.2.5
Analysis method
PCA/PVCA/Intensity boxplot/Molecule Intensity/RSD boxplot/Pearson correlation/UMAP/TSNE
Result example

notes: principal component analysis

notes: expression of metabolites in samples
Input file format requirements:
The input file supports three formats: tab delimited TXT, comma delimited CSV and XLSX. If the input file is in Excel format, when the excel file contains multiple sheets, it is required to select one sheet as the input data after uploading.
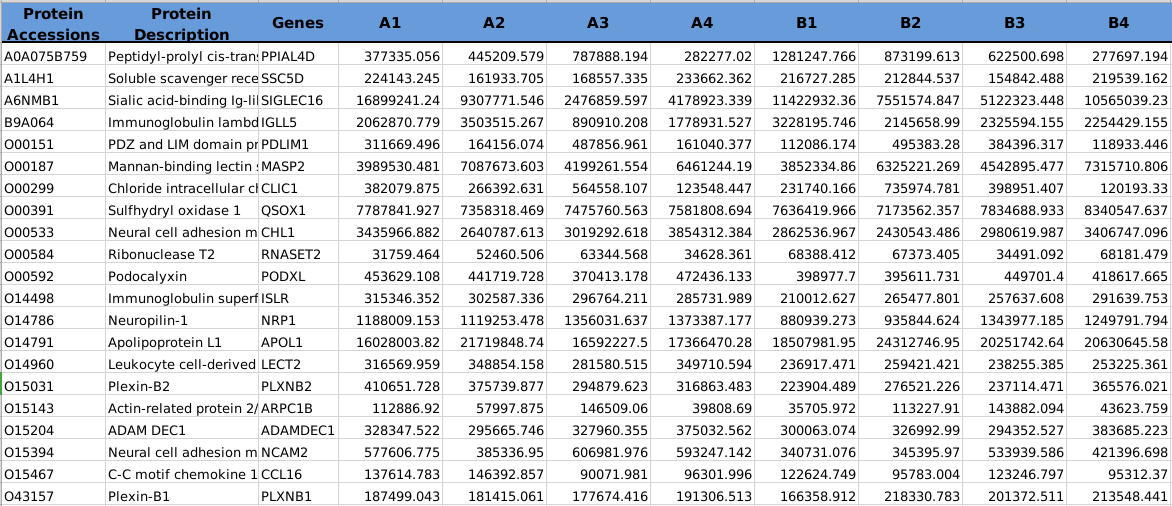
Omics data content requirements:
The column names of the table and
the first column's contents must contain non-duplicative identifiers,
such as unique protein name or metabolic name can be used as the first column.
The table should also include columns representing molecular expression data, with headers serving as sample identifiers. Including additional information is permitted,
as it will not interfere with the data analysis process.
Omics data format example:
Group design content requirements:
Group design must have ID, Type, Batch and Order columns. Pay attention to the capitalization of column names!
'ID' is the sample name contained in Omics Data;
'Type' is the grouping for sample matching;
'Batch' is a batch grouping of samples;
'Order' is the injection sequence
If the SERRF normalization method is chosen, 'Type' must include QC, and there must be at least 6 QCs in each batch.
Group design format example: